Abstract
A great challenge for early stage patients with chronic lymphocytic leukemia (CLL) is to determine if and, consequently, when they will require treatment, highlighting the need for robust prognostic assessment. Today, immunogenetic and genomic features are considered essential for accurate risk stratification in CLL. However, when utilizing such biomarkers, the prognosis is assessed assuming stable predictability over the disease course. Here, we assessed the risk for CLL evolution overtime by evaluating time-to-first-treatment (TTFT) for 2366 CLL patients not requiring treatment at the time of diagnosis (Binet A: n=1900, 80.3%/Binet B: n=287, 12.1%), hypothesizing that risk evolution might depend on both time and the biological characteristics of the clone. Based on the somatic hypermutation status (SHM) of the immunoglobulin heavy variable genes, 1364 (58%) and 1002 (42%) patients were classified as mutated (M-CLL) and unmutated CLL (U-CLL), respectively. Regarding genomic subgroups of CLL, we focused on high risk patients carrying aberrations within the TP53 gene (TP53 abn, deletion of chromosome 17p and/or TP53 mutations) and low risk patients exhibiting isolated deletion of chromosome 13q or normal FISH according to the Döhner hierarchical model (del(13q)/normal FISH).
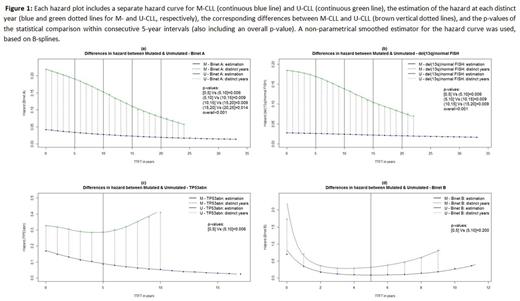
First, we estimated the risk overtime by the hazard curve. This enables to better visualize changes in risk overtime compared to a Kaplan-Meier curve by showing the estimated proportion of cases receiving treatment in a defined interval, given that a person is still treatment-free at the start of this interval. An interpolation method was then used to estimate the values of the hazard curve at each distinct year from the time of diagnosis, and the corresponding differences in hazard values between M-CLL and U-CLL. These differences constituted the basis for the overtime analysis. In particular, the overall time frame was divided in 5-year intervals, and the distributions of differences in hazard were statistically compared between consecutive 5-year intervals. P-values less than 0.05 would indicate different evolution for the hazard curve overtime for the M-CLL and U-CLL.
First, we assessed all Binet A patients, regardless of their genomic aberrations background, and found significant differences between M-CLL vs U-CLL with p[0,5]Vs(5,10]=0.006, p(5,10]Vs(10,15]=0.009, p(10,15]Vs(15,20]=0.009, p(15,20]Vs(20,25]=0.014, and p(overall)<0.001, reflecting the constant decrease of the distance between the M-CLL and U-CLL hazard curves (Fig 1a), deemed as statistically significant between consecutive 5-year intervals. Next, we focused on different genomic risk subgroups and found that del(13q)/normal FISH patients followed the M-CLL/U-CLL rule with p[0,5]Vs(5,10]=0.006, p(5,10]Vs(10,15]=0.009, p(10,15]Vs(15,20]=0.009, and p(overall)<0.001, indicating little additional impact on risk evolution beyond that dictated by the SHM status (Fig 1b). In sharp contrast, within TP53 abn patients we found p[0,5]Vs(5,10]=0.006, which reflects the statistically significant increase of the distance between the two hazard curves for M-CLL and U-CLL after the 5th year (Fig 1c), hence indicating decreasing risk for TP53 abn M-CLL and, vice versa, constantly increasing risk for TP53 abn U-CLL after the 5th year. For Binet B patients, we did not observe any significant differences with p[0,5]Vs(5,10]=0.2, highlighting a similar hazard evolution for M-CLL and U-CLL (Fig 1d). Low numbers precluded assessing the impact of genomic aberrations within this subgroup of patients.
In conclusion, within all early stage patients as well as in subgroup analysis of high or low genomic risk Binet A patients, M-CLL was found to display a rather stable overtime risk for TTFT, independently of the presence of TP53 abn. This sharply contrasted U-CLL patients, where the overtime risk for TTFT was clearly influenced by the genomic background since del(13q)/normal FISH patients showed a similar pattern with M-CLL whereas TP53 abn patients exhibited a continuously increasing risk for disease progression. From a different standpoint, these findings corroborate the concept that the precise prognostic value of TP53 abn depends on the biological background of the CLL clone that is altogether different in patients with distinct SHM status.
Strefford: Roche: Research Funding. Delgado: Abbvie, Jansen, Gilead, Roche: Consultancy, Speakers Bureau. Stavroyianni: Janssen Pharmaceuticals: Honoraria; Gilead Sciences: Honoraria; Takeda: Honoraria. Gaidano: Janssen: Consultancy, Honoraria; Gilead: Consultancy, Honoraria; AbbVie: Consultancy, Honoraria; Roche: Consultancy, Honoraria; Amgen: Consultancy, Honoraria. Ghia: AbbVie: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding; Janssen Pharmaceuticals: Honoraria, Research Funding; Roche: Honoraria; Gilead: Consultancy, Honoraria, Membership on an entity's Board of Directors or advisory committees, Research Funding, Speakers Bureau. Stamatopoulos: Abbvie: Honoraria, Research Funding; Gilead: Consultancy, Honoraria, Research Funding; Novartis SA: Research Funding; Janssen Pharmaceuticals: Honoraria, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal